- Press Office
- Press releases
- What you count is not necessarily what counts
What you count is not necessarily what counts
If scientists want to find out how fast a population of bacteria grows, they often measure how their cell count changes over time. However, this method has a major flaw: it does not measure how fast the bacteria multiply or die. Yet these factors are very important for understanding ecological processes. That is why researchers at the Max Planck Institute for Marine Microbiology in Bremen have now taken a closer look at these processes during a spring bloom in the German Bight. In doing so, they challenge some previous dogmas.
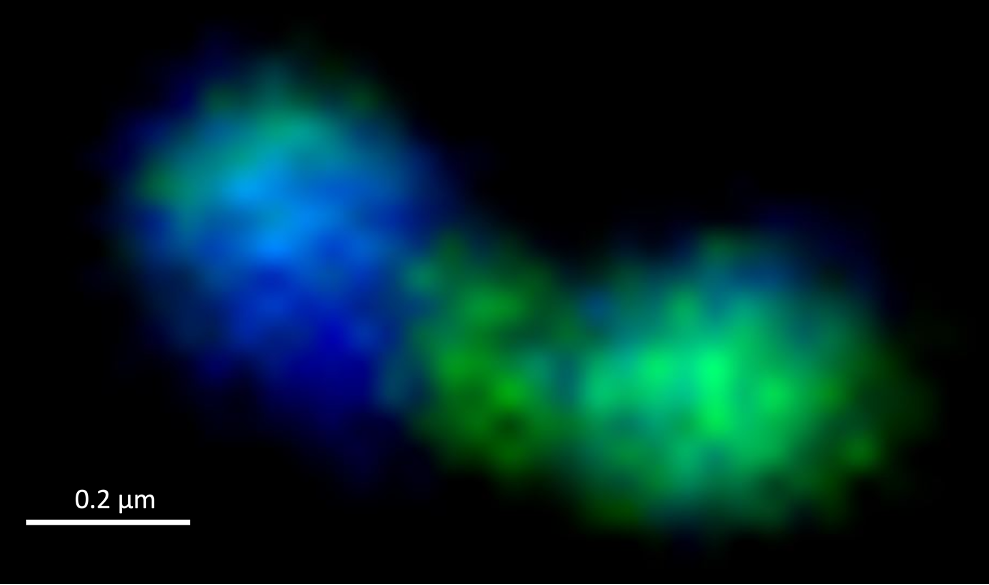
The researchers around Jan Brüwer, Bernhard Fuchs and Rudolf Amann investigated the growth of bacteria during the spring bloom off Helgoland using various methods: With the microscope, they counted and identified not only the cells present, but also the frequency of cells that were currently dividing. This way, they were able to calculate how quickly different types of bacteria multiplied in their natural environment.
“We used modern microscopic methods to visualise and count dividing cells in thousands of images,” explains Jan Brüwer, who conducted the study as part of his doctoral thesis. “We made use of the fact that a dividing cell has to split its duplicated genome into its daughter cells. Thus, we were able to clearly identify these cells based on the DNA distribution in the cell.” This enabled the researchers to determine the growth rates of individual groups of bacteria over longer periods of time.
“The results had some surprises in store for us,” says group leader Bernhard Fuchs. “For example, we found that the most common group of bacteria in the ocean, called SAR11, divides almost ten times faster than assumed.” Moreover, in many cases the measured growth rates do not match the abundance of the respective bacteria in the water. “If bacteria divide often but are not abundant, it suggests that they are a popular victim of predators or viruses,” Brüwer explains. “The timing of bacterial proliferation was also surprising: SAR11 bacteria frequently divided before the onset of the algal bloom in the North Sea. From where they took the required energy to do so is still a mystery.”
Not all bacterial groups behaved as unexpectedly as SAR11; for other groups, the results now collected were more in line with the researchers' expectations – in their case, growth rates and cell numbers largely matched.
Until now, it has been assumed that SAR11, which have very small cells, get by with little nutrients, do not divide very often and are eaten only rarely because of their small size. In contrast, other larger bacteria, for example the Bacteroidetes, are seen as popular food, multiplying quickly and disappearing just as quickly when predators and viruses get on their trail. The new study by Brüwer and his colleagues paints a very different picture.
“Our results influence our understanding of element cycles, especially the carbon cycle, in the ocean,” Brüwer emphasises. “The most abundant bacteria in the ocean, SAR11, are more active and divide faster than previously believed. This could mean that they need fewer nutrients and are a more popular food source for other organisms than suspected. Also, the general turnover of bacteria during algal blooms seems to be faster than we thought.”
“This research was methodologically very demanding and it shows how much information you can draw from microscopy images,” stresses Rudolf Amann, director at the Max Planck Institute for Marine Microbiology. “I am very proud of the researchers involved for mastering this mammoth task and glad to have the privilege to work with them. The results achieved will trigger many exciting discussions about the ecological relationships during a spring bloom and in the ocean in general.”
Original publication
Brüwer, JD, Orellana, LH, Sidhu, C, Klip, HCL, Meunier, CL, Boersma, M, Wiltshire, KH, Amann, R, Fuchs, BM (2023) In situ cell division and mortality rates of SAR11, SAR86, Bacteroidetes, and Aurantivirga during phytoplankton blooms reveal differences in population controls, mSystems (17 May 2023)
Participating institutions
- Max Planck Institute for Marine Microbiology, Celsiusstraße 1, 28359 Bremen, Germany
- Alfred Wegener Institute – Helmholtz Centre for Polar and Marine Research, Helgoland/List on Sylt, Germany
- University of Bremen, 28359 Bremen, Germany
Please direct your queries to:
Group Leader
MPI for Marine Microbiology
Celsiusstr. 1
D-28359 Bremen
Germany
|
Room: |
2222 |
|
Phone: |

Head of Press & Communications
MPI for Marine Microbiology
Celsiusstr. 1
D-28359 Bremen
Germany
|
Room: |
1345 |
|
Phone: |