- Departments
- Department of Molecular Ecology
- Integrative Microbial Genomics
Integrative Microbial Genomics
Group Leader
ERC Research Group for Ecological Genomics
Dr. Luis Humberto Orellana Retamal
MPI for Marine Microbiology
Celsiusstr. 1
D-28359 Bremen
Germany
|
Room: |
2226 |
|
Phone: |

Our Research
Our work is at the interphase between the genomic, ecological, and evolutionary significance of microbial populations. We seek to harness the power of multi-omic and molecular biology techniques to disentangle the genetic potential of natural microbial communities and link it to mechanisms involved in biochemical cycles. We are also interested in the role of overlooked microbial diversity within microbial populations in nature from a single-cell level to their ecological niche.
Method development in genomic and metagenomic approaches
We develop methodologies and approaches to precisely and effectively use (meta)genomic data. We generate approaches for analyzing unassembled short-reads when the assembly is deficient for highly complex samples. We also advance the approaches for recovering microbial genomes at the species levels using short- and long-read technologies.
Discoveries in microbial ecology using culture-independent approaches
We use computational and visualization approaches to understand the role of microbial communities. At the same time, the knowledge we gain using these high-throughput methods also helps us to generate new hypotheses that we can later test using the natural systems we study. The iterative integration of multi-omic and microbial activity is crucial in order to generate hypotheses to better understand the impact of microbes in biogeochemical cycles.
Our latest projects
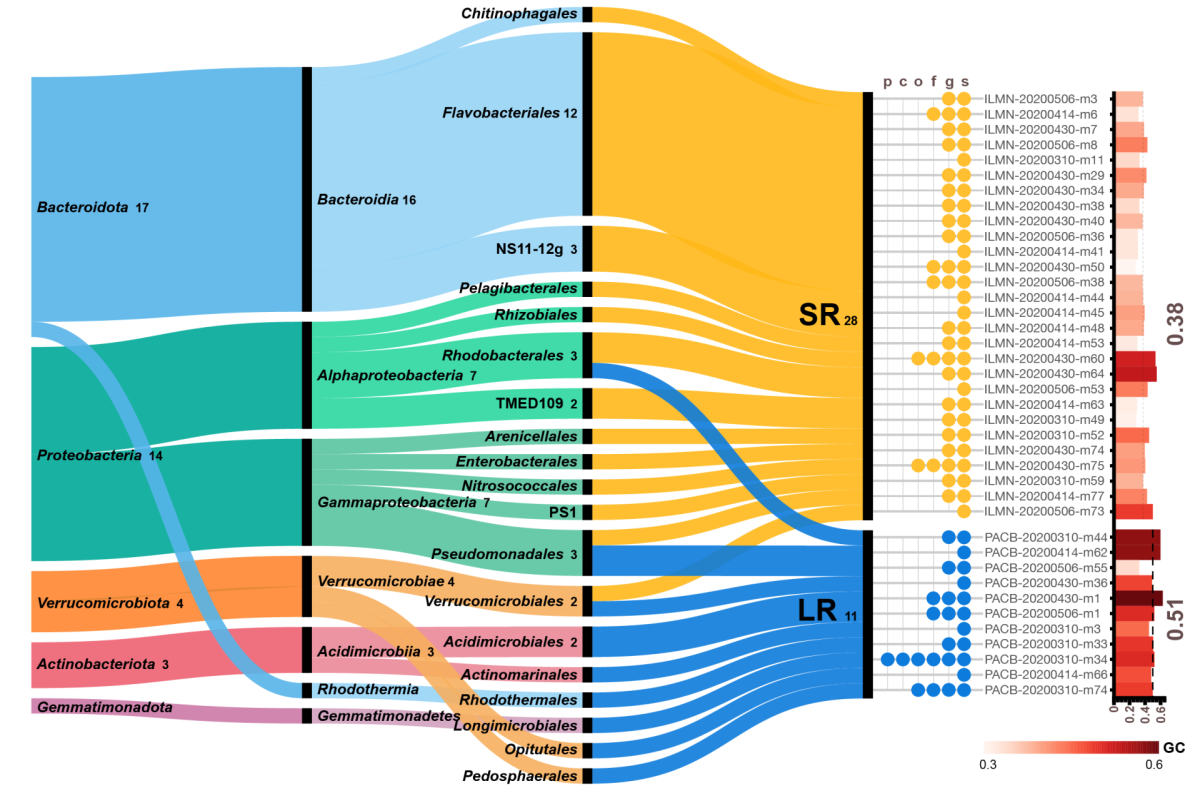
Comparing genomes recovered from time-series metagenomes using long-and short-read sequencing technologies
We compared metagenome-assembled genomes (MAGs) of the same species using short- (SR) and long-read (LR) sequencing recovered from the samples. As expected, LR MAGs were less fragmented and slightly longer than those recovered from SR metagenomic samples. Most importantly, 89% LR MAGs carried a full-length 16S rRNA (only 23% in SR MGs). We also found unique species recovered using each technology. When we examined the presence of these unique MAGs in the opposite sequencing technology, they had low sequencing depth and low breadth of coverage. Interestingly, these unique species had a GC bias depending on the sequencing platform used: unique species in LR had higher GC, whereas and those recovered from SR had lower GC. Overall, this work highlights the benefits and pitfalls when using SR and LR technologies to recover MAGs from environmental samples.
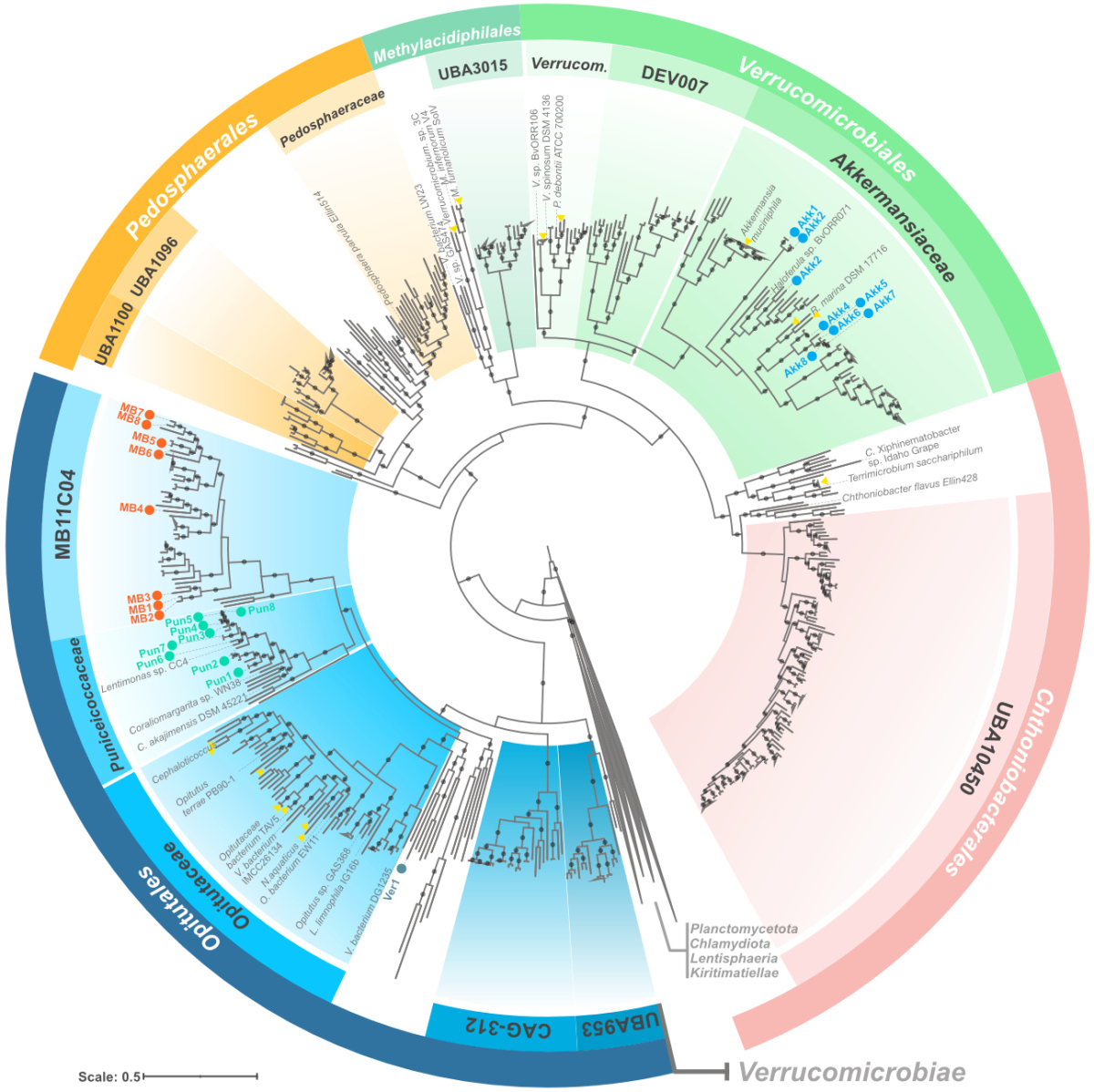
Verrucomicrobiota are specialist consumers of sulfated methyl pentoses during diatom blooms
Members of the phylum Verrucomicrobiota are enigmatic, often understudied, and underrepresented in databases. Nonetheless, their presence in different environments, from the ocean to the gut, reflects, for the moment, an untold evolutionary history of adaptations and specialization events.
Unlike other heterotrophic bacteria from the spring blooms, Verrucomicrobiota cells are well equipped to degrade highly sulfated polysaccharides containing fucose and rhamnose. Using metagenomics, metaproteomics, and cell visualization approaches, we found that Verrucomicrobiota use a combination of specialized pathways for fucose degradation that, complemented with bacterial microcompartments, can avoid the accumulation of toxic intermediates.
Suppose you were to compare the metabolic potential lightly: Verrucomicrobiota populations and prolific heterotrophs, such as members of the phylum Bacteroidota, have an equivalent number of sulfates and glycoside hydrolase genes for polysaccharide degradation. However, it’s more than just numbers: Most Verrucomicrobiota cells carry specialized degradation pathways that, linked to bacterial microcompartments, can circumvent the accumulation of the toxic lactaldehyde. No other group in the bloom unites such specialized mechanisms for the degradation of fucose and rhamnose.
Combining metagenomic approaches with CARD-FISH also allowed us to name two representative species as Candidatus Mariakkermansia forsetii and Candidatus Fucivorax forsetii. It seems you CAN make a living being picky (and specialized) at what you eat, or at least during the spring blooms.
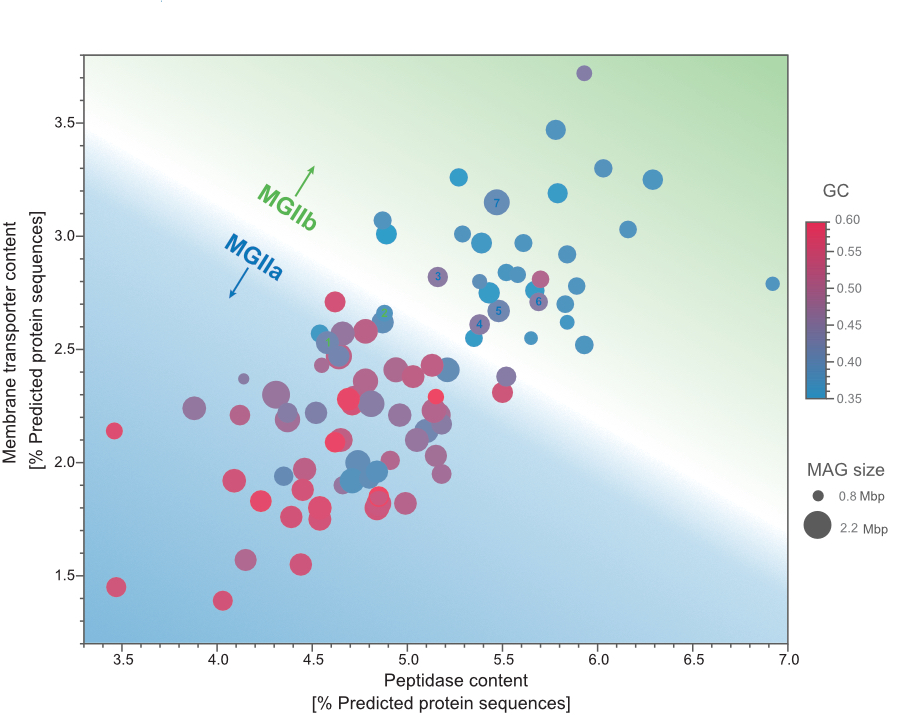
Niche differentiation among annually recurrent coastal Marine Group II Euryarchaeota
We combined metagenomic and visualization approaches to explore the Euryarchaeota MGIIa and MGIIb populations' temporal distribution in the North Sea. The combination of oligotyping and FISH approaches revealed that MGII cells were mostly free-living and of a small coccoid shape, likely resulting in grazing avoidance. Our metagenomic approach allowed us to recover MAGs that revealed differences in the genome size and GC content between MGIIa and MGIIb. Additionally, we observed highly identical and annually recurrent discrete populations for both families during the winter and summer periods hypothesize that a combination of size and metabolic potential delineates MGII populations' niches. Read more about it in the "behind the paper" blog post.
Read the paper
Team
ERC Research Group for Ecological Genomics
MPI for Marine Microbiology
Celsiusstr. 1
D-28359 Bremen
Germany
|
Room: |
3225 |
|
Phone: |

ERC Research Group for Ecological Genomics
MPI for Marine Microbiology
Celsiusstr. 1
D-28359 Bremen
Germany
|
Room: |
3225 |
|
Phone: |
