- Press Office
- Press releases 2022
- Predatory bacteria
Predatory bacteria
Most people think of microorganisms as a cause of disease rather than as its victims. But in fact, they too can become victims of bacteria that make them sick and even devour them. Such a predatory bacterium has now been described by researcher Jens Harder and his team from the Max Planck Institute for Marine Microbiology in Bremen, Germany.
The assailant: Undercover for a long time
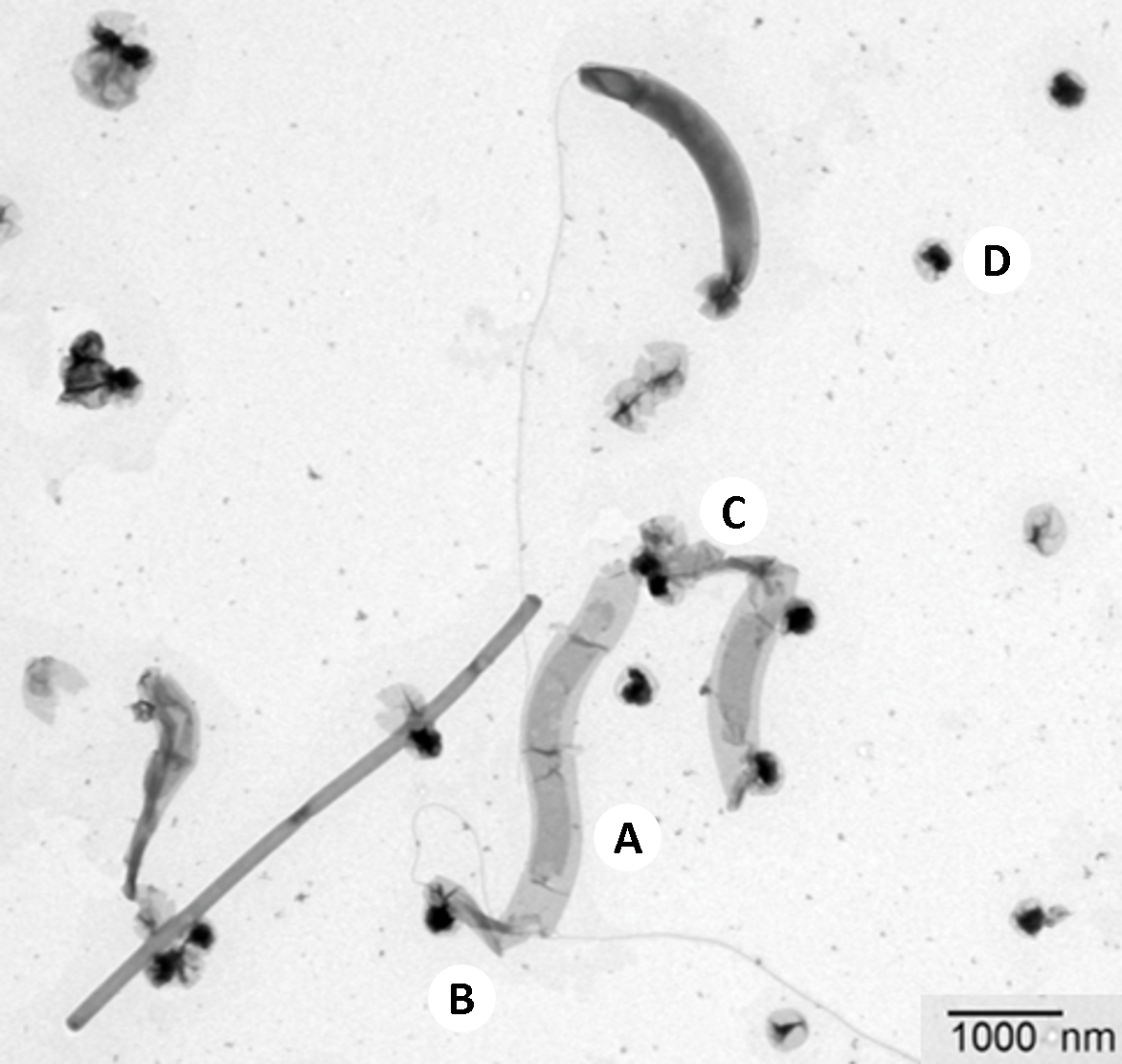
For more than twenty years, the predatory bacteria have been living largely unnoticed in Jens Harder's laboratory at the Max Planck Institute in Bremen in a so-called enrichment culture that uses limonene for methane production. They originally came from the digestion tower of the sewage treatment plant in Osterholz-Scharmbeck, close to Bremen. “We have named the new microbe Velamenicoccus archaeovorus,” says Harder. “It is an ultramicrobacterium –a particularly tiny member of the microbial world, only 200 to 300 nanometres in size and thus invisible under a normal microscope.” By comparison, a human is almost two billion nanometres tall. A few secrets of these tiny bacteria have now been revealed.
The victim: Important for biogas production
The second main character of this story, the victim, is also found in sewage treatment plants: Methanosaeta, one of the most common microbes in the world, plays a crucial role therein. This archaeon is mainly responsible for biogas production in sewage treatment plants. Individual cells of Methanosaeta live together in a protective tube, a filament. Using special dyes and a special microscope, Harder and his team were now able to prove that single cells in the Methanosaeta-filaments were sick or dead. They were limp and contained neither ribosomal nucleic acids nor genetic material – typical components of living microbial cells. The cells had presumably fallen victim to the ultramicrobacteria: “Most probably, the cause of disease is attached bacteria, and these attached bacteria are Velamenicoccus archaeovorus,” Harder explains.
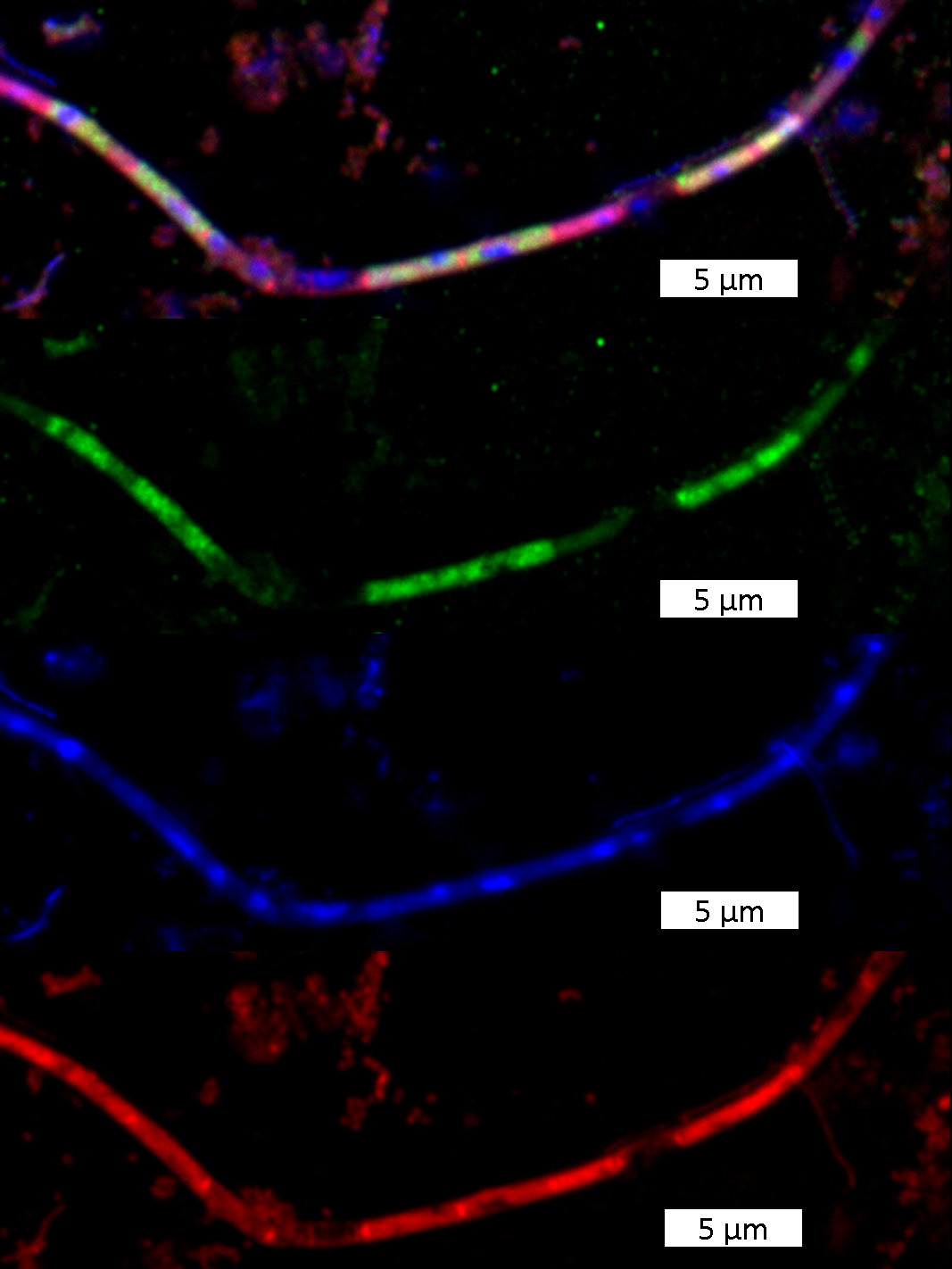
The weapon? A giant protein
“Velamenicoccus archaeovorus is not an unknown,” Harder continues. “We have found parts of its genetic material in deep sediments and other oxygen-free habitats. But what it does there was not known.” Now, researchers at the Max Planck Institute for Marine Microbiology have been able to decode the genome of this ultramicrobacterium and identify its proteins, thereby unlocking some of the tiny predator's secrets. One particularly remarkable gene is a strikingly large one. “While proteins on average consist of 333 amino acids, this gene encodes a protein with 39678 amino acids,” Harder explains. Thereby, it would be one of the largest known proteins. It is integrated into the cell wall and its surface contains enzyme domains that enable it to dissolve cells. Thus, this could well be the deadly secret of Velamenicoccus.
Ecologically significant in deep sediments?
Realising that we are dealing with such a “dangerous” bacterium permits a new look at an ecological question: Sediments are full of microorganisms, decreasing in number with increasing depth. The deeper you go, the fewer cells you will find. Until now, it was assumed that this was the result of the ongoing die-off of cells. Now another possibility arises: Microorganisms use other microorganisms as a food source, and because this is not particularly efficient, the organic material is being increasingly lost as methane and carbon dioxide. “Ultramicrobacteria can thus play a decisive role in the conversion and recycling of biomass in sediments and cause an overall reduction in biomass with depth,” Harder concludes.
Fittingly: Velamenicoccus archaeovorus, the archaea-eating microbe, belongs to the candidate phylum Omnitrophica, meaning the “all-eaters”. The finding that they live as predators now shows for the first time that this appellation is really accurate.
Original publication
Jana Kizina, Sebastian F. A. Jordan, Gerrit Alexander Martens, Almud Lonsing, Christina Probian, Androniki Kolovou, Rachel Santarella-Mellwig, Erhard Rhiel, Sten Littmann, Stephanie Markert, Kurt Stüber, Michael Richter, Thomas Schweder & Jens Harder (2022): Methanosaeta and Candidatus Velamenicoccus archaeovorus. Applied and Environmental Microbiology (ahead of print 21.03.2022).
DOI: 10.1128/aem.02407-21
Participating institutions
Max Planck Institute for Marine Microbiology, Bremen, Germany
Electron Microscopy Core Facility, EMBL Heidelberg, Heidelberg, Germany
Institute of Marine Chemistry and Biology, Carl von Ossietzky University Oldenburg, Oldenburg, Germany
Department of Pharmaceutical Biotechnology, Institute of Pharmacy, University of Greifswald, Germany
Max Planck Genome centre Cologne, Cologne, Germany
Questions?
Project Leader
Department of Molecular Ecology
MPI for Marine Microbiology
Celsiusstr. 1
D-28359 Bremen
Germany
|
Room: |
2125 |
|
Phone: |

Head of Press & Communications
MPI for Marine Microbiology
Celsiusstr. 1
D-28359 Bremen
Germany
|
Room: |
1345 |
|
Phone: |
