Flow Cytometry
What is Flow Cytometry?
Generally, flow cytometry is the measurement of single cells in a water jet. At the Max Planck Institute for Marine Microbiology, we use flow cytometry routinely to quantify cell numbers in picoplankton samples and cell cultures. Fluorescent pigments like chlorophyll and carotenoids characteristic for marine cyanobacteria and microalgae are detected as well as fluorescent dyes specific for DNA, proteins and other cellular compounds of microorganisms like cell membranes. Further, cells meeting predefined properties can be sorted with high speed in high purity for further molecular biological analysis.
Flow cytometry complements the molecular biological toolbox to study microorganisms. Its capacity of fast analysis of thousands of cells per second and the simultaneous recording of multiple parameters makes flow cytometry a standard tool in plankton research. In addition flow cytometric cell sorting (FACS) enables the physical separation and enrichment of well-defined cell populations without prior cultivation.
At the MPI in Bremen, Flow Cytometry is a research group. In this group we continuously improve our flow cytometry methods and combine them with new molecular biological approaches to gain deeper insights into the ecological role of microbes in a range of diverse marine habitats.
How does Flow Cytometry work?
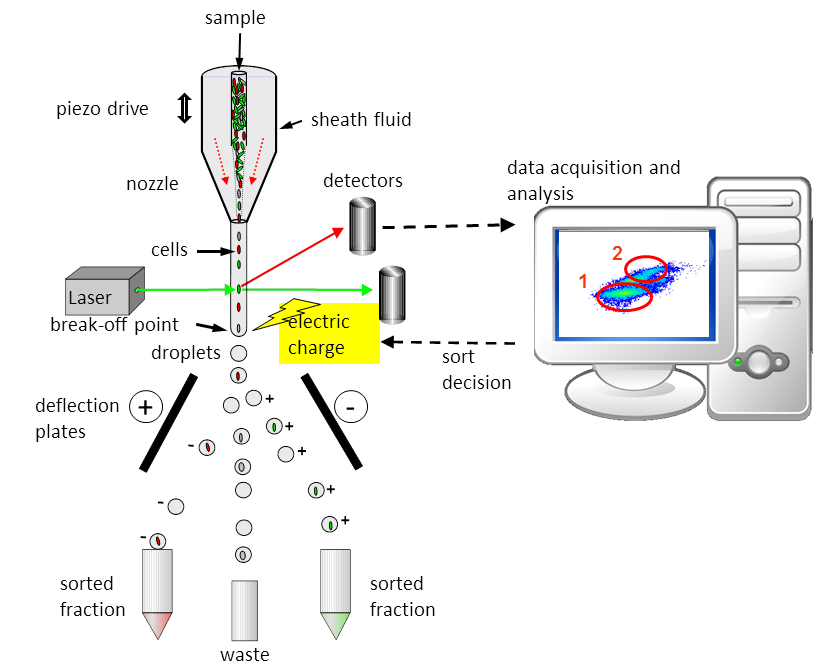
In flow cytometry, the liquid sample containing the cells is injected into the middle of a continuously flowing water jet (also called sheath fluid). The water jet is then tapered to a diameter of less than 1/10 of a millimetre using a nozzle. The enclosed cells are thus focused into the centre of the water jet and exit the nozzle like beads on a string. The precisely aligned cells pass the laser beam individually at the observation point and can thus be excited by the laser light. Depending on the size and staining of the cell, the light signals from the cells are focused through lenses (not shown) onto the detectors, where they are converted into electrical signals and further fed into software on the computer.
In sorting mode, the cells can also be separated according to certain properties. To do this, the researchers set conditions that the cells must fulfil. The nozzle oscillates in high-frequency oscillations, which causes the water jet to break off into defined droplets below the observation point. If a cell that meets the specified parameters is detected, the corresponding cell is electrically charged with its droplet. The charged droplets with the cells are deflected according to their charge (positive or negative) and land in a reaction vessel. Shown here is a two-way sorter that can sort two populations simultaneously.
We currently have three different flow cytometers at the Institute. They differ for example in size, sensitivity, and capability. All three can be transported and are suitable for use on board research vessels. Further information is available on the department page of the Flow Cytometry Group.
Flow Cytometry in action
Analysis of unknown microbes
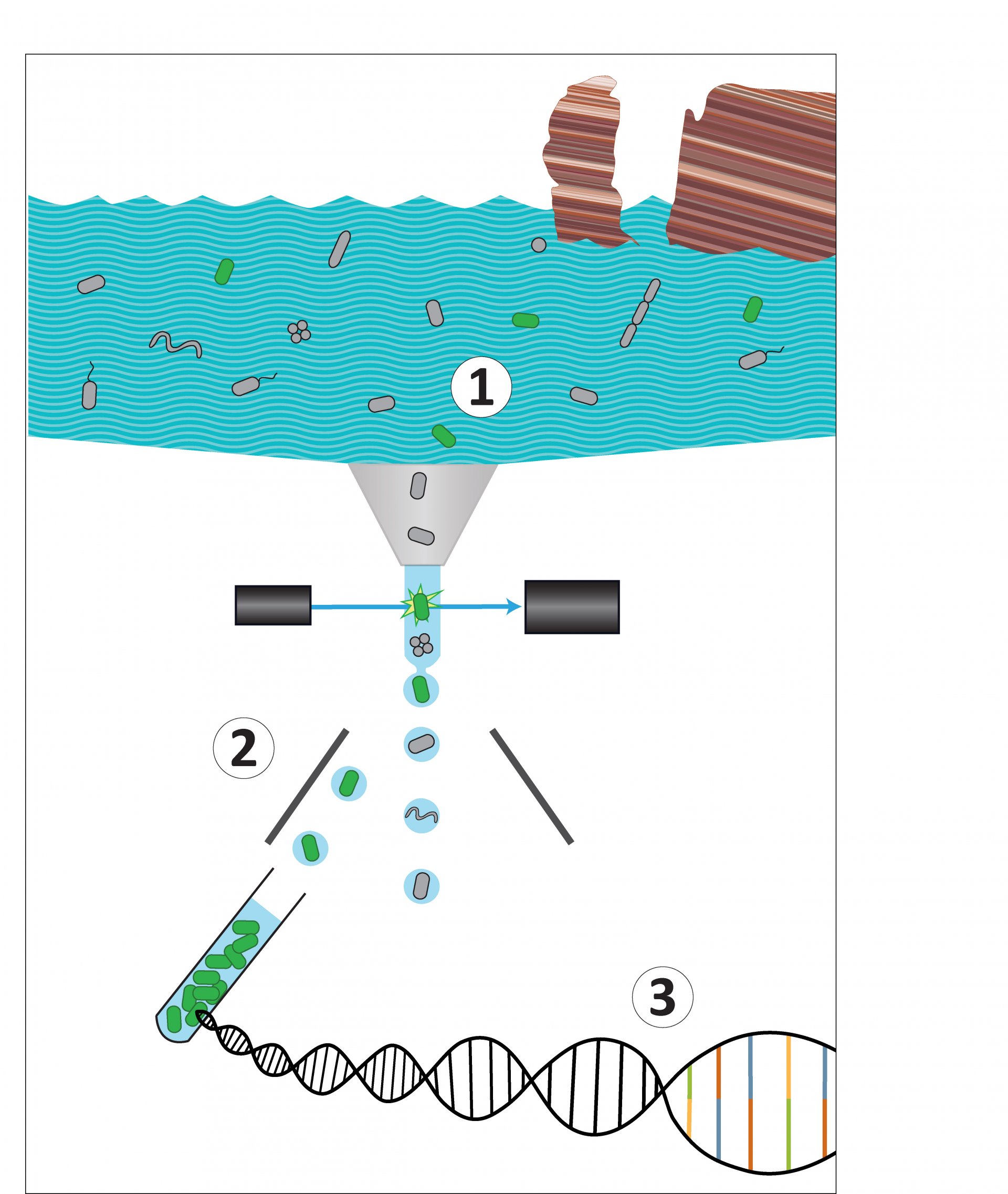
Marine microbes like to play hide and seek. Some bacteria often appear in samples but do not grow in the lab. At the same time, they are too few to be discovered via genetic analysis.
The idea to solve this problem: As we have a flow cytometer with a cell sorting system in our research group, the idea arised to select the cells before sequencing the DNA. That way, the diversity is reduced and the rare species can no longer hide. Thus, the scientists used the FISH-method to label DNA sections of the microbes with fluorescent dyes to detect and sort the bacteria they were interested in. However, they faced two challenges: Firstly, a very bright FISH signal was needed for the detection and sorting of targeted cells using flow cytometry. Secondly, sufficient unimpaired DNA material was required for high quality genome sequencing.
A team around the scientist Anissa Grieb, together with researchers from the Joint Genome Institute, tested how this could work. At the end, they succeeded by using a recently developed version of the FISH-method, the hybridization chain reaction (HCR)-FISH. First, they optimized this procedure with pure cultures in the laboratory and afterwards used it on the environmental samples, which had been taken off the coast of Helgoland.
- You can find more information about this method in the press release "Analysis of unknown microbes is getting easier"
- Here you find the paper: Grieb A, Bower RM, Oggerin M, Goudeau D, Lee J, Malmstrom RR, Woyke T, Fuchs BM. 2020. A pipeline for targeted metagenomics of environmental bacteria. Microbiome 8:21 https://doi.org/10.1186/s40168-020-0790-7
Who uses Flow Cytometry?
It is mainly used by scientists of the department Molecular Ecology. However, it is also open to all other scientists of the institute as well as to external researchers in the context of collaborative projects.
Contact
Group Leader
MPI for Marine Microbiology
Celsiusstr. 1
D-28359 Bremen
Germany
|
Room: |
2222 |
|
Phone: |