- Departments
- Department of Molecular Ecology
- Helgoland Roads
Helgoland Roads
Project Leader
Project Leader
Department of Molecular Ecology
MPI for Marine Microbiology
Celsiusstr. 1
D-28359 Bremen
Germany
|
Room: |
2223 |
|
Phone: |

Spring blooms at Helgoland
Blooms of marine algae are a global phenomenon, in particular in nutrient-rich coastal areas during spring. After winter with increasing sunlight, diatoms and other uni- to pluricellular microalgae start to proliferate. Since usually only a few predators are present during these times, the algae can proliferate very fast. The resulting algal blooms can cover such large areas that they are often visible via satellite imagery from space. It is estimated that marine algal growth is responsible for about half of the annual photosynthetic carbon dioxide fixation, and thus algae play a crucial role in global carbon cycling.
In Germany, the island Helgoland is among the most well-suited sites to study algae blooms. Helgoland is located in the German Bight (southern North Sea) and is Germany’s only off-shore island. Together with our collaboration partners from the University of Greifswald and the Alfred Wegener Institute in Bremerhaven, we have been studying spring blooms intensively since 2009 (Teeling, Fuchs et al., 2009).
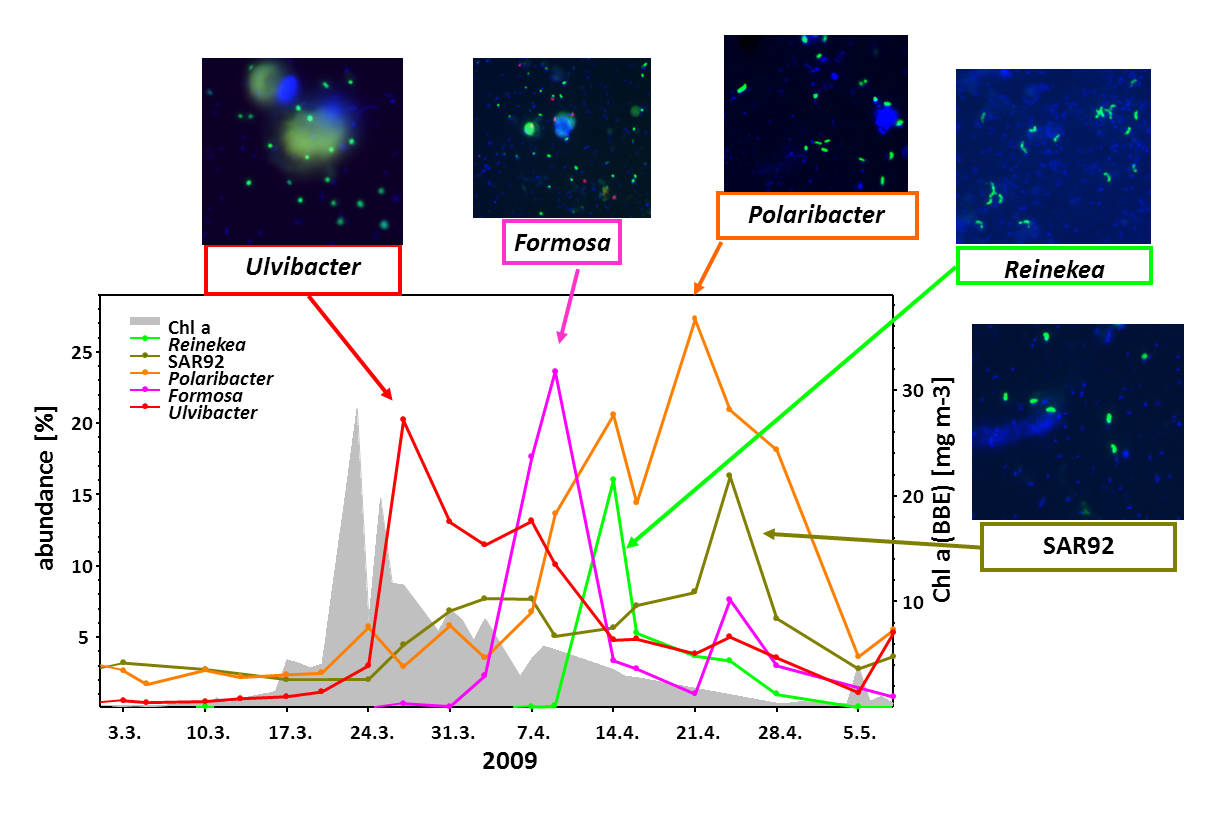
While algae blooms tend to differ from year to year, there are recurrent patterns. At first, nutrients are aplenty and algae can go into almost unrestricted growth. After a while, algal growth ceases because nutrients become limiting, grazing by protists (unicellular animals) increases, and also viruses attack the algae. Ultimately, the blooms collapse.
During algae blooms and in particular, during the terminal bloom phases, large amounts of algal organic compounds are released into the water and thus become available to free-living marine bacteria (“bacterioplankton”). Among the major classes of algal-derived substrates besides proteins and lipids are polysaccharides. The reason for this is that algae use differently composed polysaccharides for various purposes, i.e. as cell wall constituents, as exterior slimes (what makes macro-algae feel slimy), as stabilizing matrix components, or for energy storage.
We have analyzed the bacterial response to algae blooms in great detail. In a data-rich study we could show that certain clades of responding bacteria are annually recurring (Teeling, Fuchs et al., 2016). In particular certain clades of the Bacteroidetes, such as members of the genus Polaribacter (Avcı et al., 2020), were always present in large numbers.
Polysaccharide utilization loci
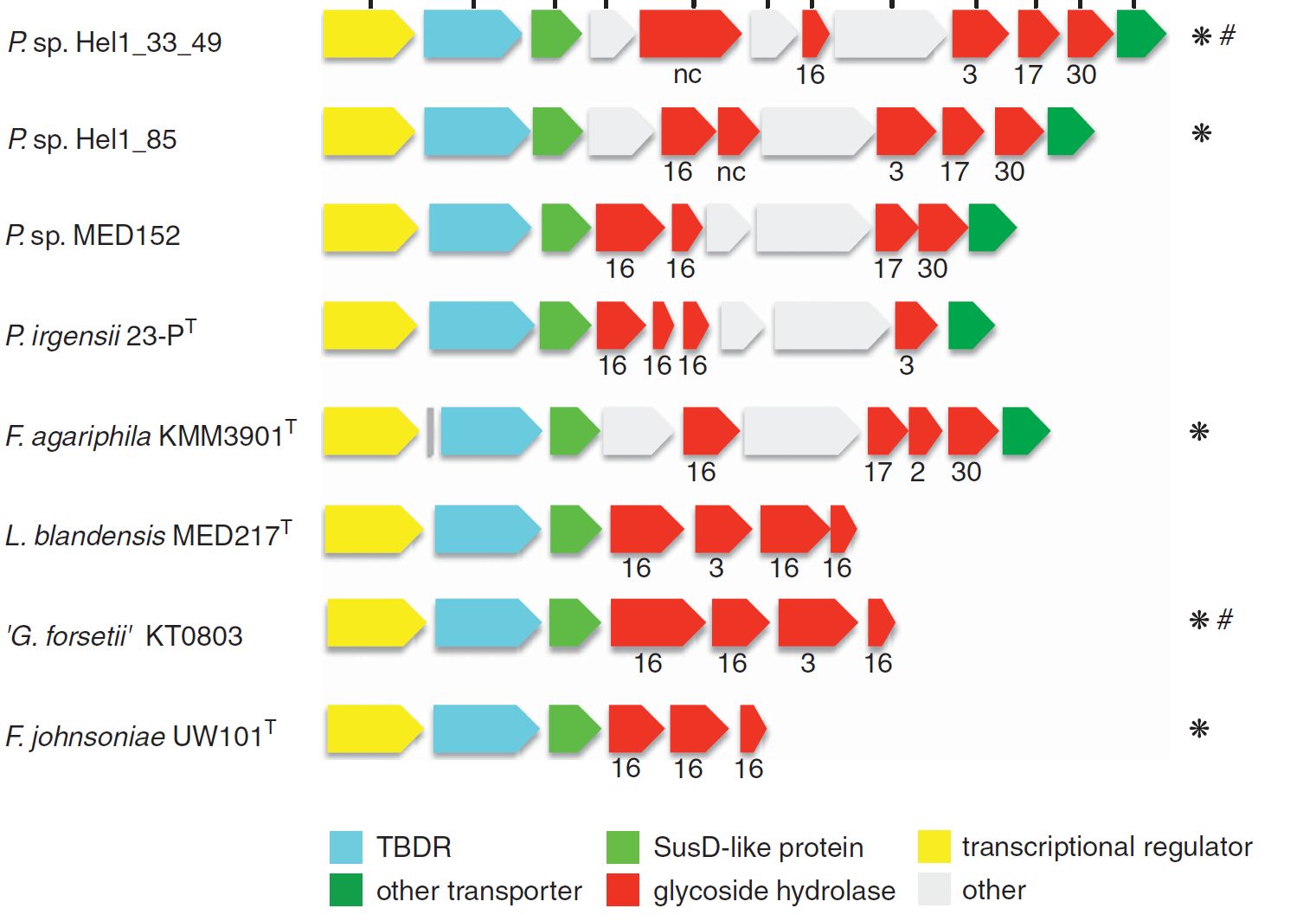
A survey of collective genomes (“metagenomes”) of bloom-associated bacteria revealed a prevalence of genes for the decomposition of algal polysaccharides in many of the abundant Bacteroidetes clades. These genes code for so-called carbohydrate-active enzymes (“CAZymes”), and they are usually clustered in polysaccharide utilization loci (PULs) of sometimes only a few and sometimes dozens of genes. Typically, a PUL has the genes for the cellular machinery to decompose one specific type of polysaccharide. Since algae produce a large number of different polysaccharides, no bacterium has the genes to decompose them all. Instead, different bacteria specialize on the decomposition of different polysaccharide types. Thus, the bacterial decomposition of algal biomass during and after algae blooms is an achievement of an entire community of specialized bacteria.
The composition of PULs can provide hints on the approximate type of polysaccharide that a given bacterium can decompose. In order to study PUL diversity within North Sea Bacteroidetes, we sequenced 53 species that were isolated from the North Sea in the group of Prof. Dr. Jens Harder (Kappelmann et al., 2016). More recently, we analyzed 38 metagenomes that were obtained at different times during spring phytoplankton blooms in 2010 to 2012 and could show that the bulk of algal polysaccharides is likely decomposed by about a dozen abundant and recurrent Bacteroidetes clades (Krüger et al., 2019). Our current focus is to study these clades via isolated representatives and via high time-resolution onsite activity measurements (e.g. metagenomics & metatranscriptomics).
Funding
This research is part of the POMPU (Proteogenomics of Marine Polysaccharide Utilization) project funded by the DFG (Deutsche Forschungsgemeinschaft; project FOR2406). It belongs to POMPU subproject B1 that is supported by DFG grants to Dr. Hanno Teeling (TE 813/2-1) and to Prof. Dr. Rudolf I. Amann (AM 73/9-1).
Current Project Members
Guest
Department of Molecular Ecology
MPI for Marine Microbiology
Celsiusstr. 1
D-28359 Bremen
Germany

PhD student
Department of Molecular Ecology
MPI for Marine Microbiology
Celsiusstr. 1
D-28359 Bremen
Germany
|
Room: |
2223 |
|
Phone: |

Associated Project Members
Dechen Lu
[Bitte aktivieren Sie Javascript]
Recent Project Members
Dr. Burak Avcı
Dr. Lennart Kappelmann
Dr. Megan Chafee
Dr. Jing Chen
Dr. Peng Xing
Dr. Sixing Huang